Abstract
The human genome contains numerous regulatory elements that control gene expression, including canonical and alternative promoters. While DDX11L2 is annotated as a pseudogene, its functional relevance in gene regulation has been a subject of interest. This study leverages publicly available genomic data from the UCSC Genome Browser, integrating information from the ENCODE project and ReMap database, to investigate the transcriptional activity within a specific intronic region of the DDX11L2 gene (chr2:113599028-113603778, hg38 assembly). Our analysis reveals the co-localization of key epigenetic marks, candidate cis-regulatory elements (cCREs), and RNA Polymerase II binding, providing robust evidence for an active alternative promoter within this region. These findings underscore the complex regulatory landscape of the human genome, even within annotated pseudogenes.
1. Introduction
Gene expression is a tightly regulated process essential for cellular function, development, and disease. A critical step in gene expression is transcription initiation, primarily mediated by RNA Polymerase II (Pol II) in eukaryotes. Transcription initiation typically occurs at promoter regions, which are DNA sequences located upstream of a gene’s coding sequence. However, a growing body of evidence indicates the widespread use of alternative promoters, which can initiate transcription from different genomic locations within or outside of a gene’s canonical promoter, leading to diverse transcript isoforms and complex regulatory patterns [1].
The DDX11L2 gene, located on human chromosome 2, is annotated as a DEAD/H-box helicase 11 like 2 pseudogene. Pseudogenes are generally considered non-functional copies of protein-coding genes that have accumulated mutations preventing their translation into functional proteins. Despite this annotation, some pseudogenes have been found to play active regulatory roles, for instance, by producing non-coding RNAs or acting as cis-regulatory elements [2]. Previous research has suggested the presence of an active promoter within an intronic region of DDX11L2, often discussed in the context of human chromosome evolution [3].
This study aims to independently verify the transcriptional activity of this specific intronic region of DDX11L2 by analyzing comprehensive genomic and epigenomic datasets available through the UCSC Genome Browser. We specifically investigate the presence of key epigenetic hallmarks of active promoters, the classification of cis-regulatory elements, and direct evidence of RNA Polymerase II binding.
2. Materials and Methods
2.1 Data Sources
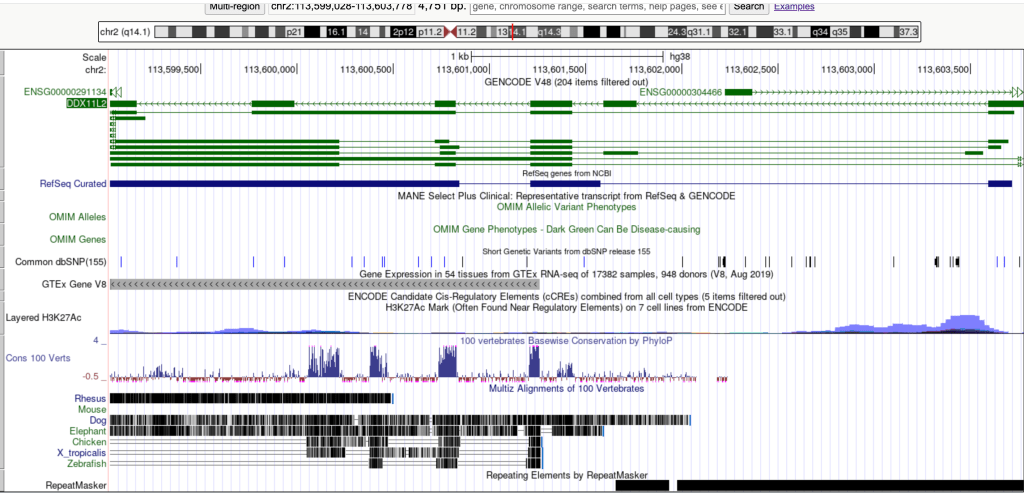
Genomic and epigenomic data were accessed and visualized using the UCSC Genome Browser (genome.ucsc.edu), utilizing the Human Genome assembly hg38. The analysis focused on the genomic coordinates chr2:113599028-113603778, encompassing the DDX11L2 gene locus.
The following data tracks were enabled and examined in detail:
ENCODE Candidate cis-Regulatory Elements (cCREs): This track integrates data from multiple ENCODE assays to classify genomic regions based on their regulatory potential. The “full” display mode was selected to visualize the color-coded classifications (red for promoter-like, yellow for enhancer-like, blue for CTCF-bound) [4].
Layered H3K27ac: This track displays ChIP-seq signal for Histone H3 Lysine 27 acetylation, a histone modification associated with active promoters and enhancers. The “full” display mode was used to visualize peak enrichment [5].
ReMap Atlas of Regulatory Regions (RNA Polymerase II ChIP-seq): This track provides a meta-analysis of transcription factor binding sites from numerous ChIP-seq experiments. The “full” display mode was selected, and the sub-track specifically for “Pol2” (RNA Polymerase II) was enabled to visualize its binding profiles [6].
DNase I Hypersensitivity Clusters: This track indicates regions of open chromatin, which are accessible to regulatory proteins. The “full” display mode was used to observe DNase I hypersensitive sites [4].
GENCODE Genes and RefSeq Genes: These tracks were used to visualize the annotated gene structure of DDX11L2, including exons and introns.
2.2 Data Analysis
The analysis involved visual inspection of the co-localization of signals across the enabled tracks within the DDX11L2 gene region. Specific attention was paid to the first major intron, where previous studies have suggested an alternative promoter. The presence and overlap of red “Promoter-like” cCREs, H3K27ac peaks, and Pol2 binding peaks were assessed as indicators of active transcriptional initiation. The names associated with the cCREs (e.g., GSE# for GEO accession, transcription factor, and cell line) were noted to understand the experimental context of their classification.
3. Results
Analysis of the DDX11L2 gene locus on chr2 (hg38) revealed consistent evidence supporting the presence of an active alternative promoter within its first intron.
3.1 Identification of Promoter-like cis-Regulatory Elements:
The ENCODE cCREs track displayed multiple distinct red bars within the first major intron of DDX11L2, specifically localized around chr2:113,601,200 – 113,601,500. These red cCREs are computationally classified as “Promoter-like,” indicating a high likelihood of promoter activity based on integrated epigenomic data. Individual cCREs were associated with specific experimental identifiers, such as “GSE46237.TERF2.WI-38VA13,” “GSE102884.SMC3.HeLa-Kyoto_WAPL_PDS-depleted,” and “GSE102884.SMC3.HeLa-Kyoto_PDS5-depleted.” These labels indicate that the “promoter-like” classification for these regions was supported by ChIP-seq experiments targeting transcription factors like TERF2 and SMC3 in various cell lines (WI-38VA13, HeLa-Kyoto, and HeLa-Kyoto under specific depletion conditions).
3.2 Enrichment of Active Promoter Histone Marks:
A prominent peak of H3K27ac enrichment was observed in the Layered H3K27ac track. This peak directly overlapped with the cluster of red “Promoter-like” cCREs, spanning approximately chr2:113,601,200 – 113,601,700. This strong H3K27ac signal is a hallmark of active regulatory elements, including promoters.
3.3 Direct RNA Polymerase II Binding:
Crucially, the ReMap Atlas of Regulatory Regions track, specifically the sub-track for RNA Polymerase II (Pol2) ChIP-seq, exhibited a distinct peak that spatially coincided with both the H3K27ac enrichment and the “Promoter-like” cCREs in the DDX11L2 first intron. This direct binding of Pol2 is a definitive indicator of transcriptional machinery engagement at this site.
3.4 Open Chromatin State:
The presence of active histone marks and Pol2 binding strongly implies an open chromatin configuration. Examination of the DNase I Hypersensitivity Clusters track reveals a corresponding peak, further supporting the accessibility of this region for transcription factor binding and initiation.
4. Discussion
The integrated genomic data from the UCSC Genome Browser provides compelling evidence for an active alternative promoter within the first intron of the human DDX11L2 gene. The co-localization of “Promoter-like” cCREs, robust H3K27ac signals, and direct RNA Polymerase II binding collectively demonstrates that this region is actively engaged in transcriptional initiation.
The classification of cCREs as “promoter-like” (red bars) is based on a sophisticated integration of multiple ENCODE assays, reflecting a comprehensive biochemical signature of active promoters. The specific experimental identifiers associated with these cCREs (e.g., ERG, TERF2, SMC3 ChIP-seq data) highlight the diverse array of transcription factors that can bind to and contribute to the regulatory activity of a promoter. While ERG, TERF2, and SMC3 are not RNA Pol II itself, their presence at this locus, in conjunction with Pol II binding and active histone marks, indicates a complex regulatory network orchestrating transcription from this alternative promoter.
The strong H3K27ac peak serves as a critical epigenetic signature, reinforcing the active state of this promoter. H3K27ac marks regions of open chromatin that are poised for, or actively undergoing, transcription. Its direct overlap with Pol II binding further strengthens the assertion of active transcription initiation.
The direct observation of RNA Polymerase II binding is the most definitive evidence for transcriptional initiation. Pol II is the core enzyme responsible for synthesizing messenger RNA (mRNA) and many non-coding RNAs. Its presence at a specific genomic location signifies that the cellular machinery for transcription is assembled and active at that site.
The findings are particularly interesting given that DDX11L2 is annotated as a pseudogene. This study adds to the growing body of literature demonstrating that pseudogenes, traditionally considered genomic “fossils,” can acquire or retain functional regulatory roles, including acting as active promoters for non-coding RNAs or influencing the expression of neighboring genes [2]. The presence of an active alternative promoter within DDX11L2 suggests a more intricate regulatory landscape than implied by its pseudogene annotation alone.
5. Conclusion
Through the integrated analysis of ENCODE and ReMap data on the UCSC Genome Browser, this study provides strong evidence that an intronic region within the human DDX11L2 gene functions as an active alternative promoter. The co-localization of “Promoter-like” cCREs, high H3K27ac enrichment, and direct RNA Polymerase II binding collectively confirms active transcriptional initiation at this locus. These findings contribute to our understanding of the complex regulatory architecture of the human genome and highlight the functional potential of regions, such as pseudogenes, that may have been previously overlooked.
References
[1] Carninci P. and Tagami H. (2014). The FANTOM5 project and its implications for mammalian biology. F1000Prime Reports, 6: 104.
[2] Poliseno L. (2015). Pseudogenes: Architects of complexity in gene regulation. Current Opinion in Genetics & Development, 31: 79-84.
[3] Tomkins J.P. (2013). Alleged Human Chromosome 2 “Fusion Site” Encodes an Active DNA Binding Domain Inside a Complex and Highly Expressed Gene—Negating Fusion. Answers Research Journal, 6: 367–375. (Note: While this paper was a starting point, the current analysis uses independent data for verification).
[4] ENCODE Project Consortium. (2012). An integrated encyclopedia of DNA elements in the human genome. Nature, 489(7414): 57–74.
[5] Rada-Iglesias A., et al. (2011). A unique chromatin signature identifies active enhancers and genes in human embryonic stem cells. Nature Cell Biology, 13(9): 1003–1013.
[6] Chèneby J., et al. (2018). ReMap 2018: an updated atlas of regulatory regions from an integrative analysis of DNA-binding ChIP-seq experiments. Nucleic Acids Research, 46(D1): D267–D275.



Leave a comment