A Creationist Perspective
The debate surrounding the origin and diversification of life continues, with proponents of neo-Darwinian evolution often citing observed instances of speciation and adaptations as evidence for macroevolution and the gradual development of complex biological systems. A recent “MEGA POST” on Reddit’s r/DebateEvolution presented several cases purported to demonstrate these processes, challenging the creationist understanding of life’s history. This article will examine these claims from a young-Earth creationist viewpoint.
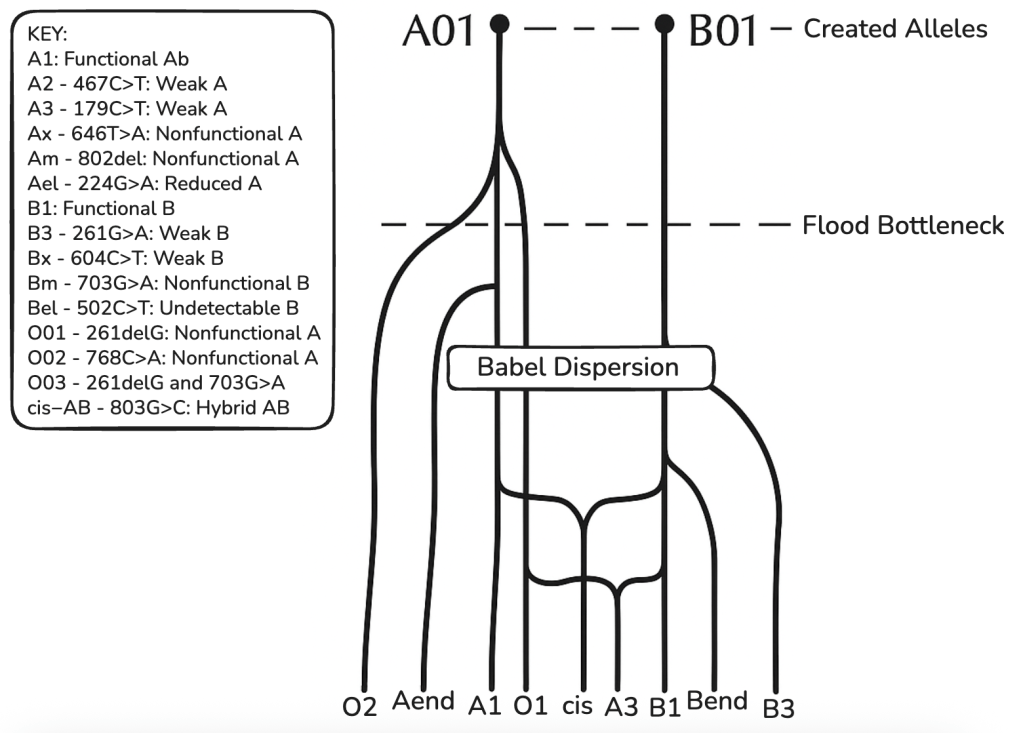
The original post defined key terms, stating, “Macroevolution ~ variations in heritable traits in populations with multiple species over time. Speciation marks the start of macroevolution.” However, creationists distinguish between microevolution – variation and speciation within a created kind – and macroevolution – the hypothetical transition between fundamentally different kinds of organisms. While the former is observable and acknowledged, the latter lacks empirical support and the necessary genetic mechanisms.
Alleged Cases of Macroevolution:
The post presented eleven cases as evidence of macroevolution.
1. Lizards evolving placentas: The observation of reproductive isolation in Zootoca vivipara with different modes of reproduction was highlighted. The author noted, “(This is probably my favourite example of the bunch, as it shows a highly non-trivial trait emerging, together with isolation, speciation and selection for the new trait to boot.)” From a creationist perspective, the development of viviparity within lizards likely involves the expression or modification of pre-existing genetic information within the lizard kind. This adaptation and speciation do not necessitate the creation of novel genetic information required for a transition to a different kind of organism.
2. Fruit flies feeding on apples: The divergence of the apple maggot fly (Rhagoletis pomonella) into host-specific groups was cited as sympatric speciation. This adaptation to different host plants and the resulting reproductive isolation are seen as microevolutionary changes within the fruit fly kind, utilizing the inherent genetic variability.
3. London Underground mosquito: The adaptation of Culex pipiens f. molestus to underground environments was presented as allopatric speciation. The observed physiological and behavioral differences, along with reproductive isolation, are consistent with diversification within the mosquito kind due to environmental pressures acting on the existing gene pool.
4. Multicellularity in Green Algae: The lab observation of obligate multicellularity in Chlamydomonas reinhardtii under predation pressure was noted. The author stated this lays “the groundwork for de novo multicellularity.” While this is an interesting example of adaptation, the transition from simple coloniality to complex, differentiated multicellularity, as seen in plants and animals, requires a significant increase in genetic information and novel developmental pathways. The presence of similar genes across different groups could point to a common designer employing similar modules for diverse functions.
5. Darwin’s Finches, revisited 150 years later: Speciation in the “Big Bird lineage” due to environmental pressures was discussed. This classic example of adaptation and speciation on the Galapagos Islands demonstrates microevolutionary changes within the finch kind, driven by natural selection acting on existing variations in beak morphology.
6 & 7. Salamanders and Greenish Warblers as ring species: These examples of geographic variation leading to reproductive isolation were presented as evidence of speciation. While ring species illustrate gradual divergence, the observed changes occur within the salamander and warbler kinds, respectively, and do not represent transitions to fundamentally different organisms.
8. Hybrid plants and polyploidy: The formation of Tragopogon miscellus through polyploidy was cited as rapid speciation. The author noted that crossbreeding “exploits polyploidy…to enhance susceptibility to selection for desired traits.” Polyploidy involves the duplication of existing chromosomes and the combination of genetic material from closely related species within the plant kingdom. This mechanism facilitates rapid diversification but does not generate the novel genetic information required for macroevolutionary transitions.
9. Crocodiles and chickens growing feathers: The manipulation of gene expression leading to feather development in these animals was discussed. The author suggested this shows “how birds are indeed dinosaurs and descend within Sauropsida.” Creationists interpret the shared genetic toolkit and potential for feather development within reptiles and birds as evidence of a common design within a broader created kind, rather than a direct evolutionary descent in the Darwinian sense.
10. Endosymbiosis in an amoeba: The observation of a bacterium becoming endosymbiotic within an amoeba was presented as analogous to the origin of organelles. Creationists propose that organelles were created in situ with their host cells, designed for symbiotic relationships from the beginning. The observed integration is seen as a function of this initial design.
11. Eurasian Blackcap: The divergence in migratory behavior and morphology leading towards speciation was highlighted. This represents microevolutionary adaptation within the bird kind in response to environmental changes.
Addressing “Irreducible Complexity”:
The original post also addressed the concept of irreducible complexity with five counter-examples.
1. E. Coli Citrate Metabolism in the LTEE: The evolution of citrate metabolism was presented as a refutation of irreducible complexity. The author noted that this involved “gene duplication, and the duplicate was inserted downstream of an aerobically-active promoter.” While this demonstrates the emergence of a new function, it occurred within the bacterial kind and involved the modification and duplication of existing genetic material. Therefore, is no evidence here to suggest an evolutionary pathway for the origin of citrate metabolism.
2. Tetherin antagonism in HIV groups M and O: The different evolutionary pathways for overcoming tetherin resistance were discussed. Viruses, with their rapid mutation rates and unique genetic mechanisms, present a different case study than complex cellular organisms. This is not analogous in the slightest.
3. Human lactose tolerance: The evolution of lactase persistence was presented as a change that is “not a loss of regulation or function.” This involves a regulatory mutation affecting the expression of an existing gene within the human genome. Therefore, it’s not a gain either. This is just a semantic game.
4. Re-evolution of bacterial flagella: The substitution of a key regulatory protein for flagellum synthesis was cited. The author noted this is “an incredibly reliable two-step process.” While this demonstrates the adaptability of bacterial systems, the flagellum itself remains a complex structure with numerous interacting components – none of said components have gained or lost the cumulative necessary functions.
5. Ecological succession: The development of interdependent ecosystems was presented as a challenge to irreducible complexity. However, ecological succession describes the interactions and development of communities of existing organisms, not the origin of the complex biological systems within those organisms.
Conclusion:
While the presented cases offer compelling examples of adaptation and speciation, we interpret these observations as occurring within the boundaries of created kinds, utilizing the inherent genetic variability designed within them. These examples do not provide conclusive evidence for macroevolution – the transition between fundamentally different kinds of organisms – nor do they definitively refute the concept of irreducible complexity in the origin of certain biological systems. The fact that so many of these are, if not neutral, loss-of-function or loss-of-information mutations creates a compelling case for creation as the inference to the best explanation. The creationist model, grounded in the historical robustness of the Biblical account and supported by scientific evidence (multiple cross-disciplinary lines), offers a coherent alternative explanation for the diversity and complexity of life. As the original post concluded,
“if your only response to the cases of macroevolution are ‘it’s still a lizard’, ‘it’s still a fly you idiot’ etc, congrats, you have 1) sorely missed the point and 2) become an evolutionist now!”
However, the point is not that change doesn’t occur (we expect that on our model), but rather the kind and extent of that change, which, from a creationist perspective, remains within divinely established explanatory boundaries of the creation model and contradicts a universal common descent model.
References:
Teixeira, F., et al. (2017). The evolution of reproductive isolation during a rapid adaptive radiation in alpine lizards. Proceedings of the National Academy of Sciences, 114(12), E2386-E2393. https://doi.org/10.1073/pnas.1635049100
Fonseca, D. M., et al. (2023). Rapid Speciation of the London Underground Mosquito Culex pipiens molestus. ResearchGate. https://doi.org/10.13140/RG.2.2.23813.22247
Grant, P. R., & Grant, B. R. (2017). Texas A&M professor’s study of Darwin’s finches reveals species can evolve in two generations. Texas A&M Today. https://stories.tamu.edu/news/2017/12/01/texas-am-professors-study-of-darwins-finches-reveals-species-can-evolve-in-two-generations/
Feder, J. L., et al. (1997). Allopatric host race formation in sympatric hawthorn maggot flies. Proceedings of the National Academy of Sciences, 94(15), 7761-7766. https://doi.org/10.1073/pnas.94.15.7761
Tishkoff, S. A., et al. (2013). Convergent adaptation of human lactase persistence in Africa and Europe. Nature Genetics, 45(3), 233-240. https://doi.org/10.1038/ng.2529 (Note: While the URL provided redirects to PMC, the original publication is in Nature Genetics. I have cited the primary source.)